Simulation Metadata Specification
PLEASE NOTE: development of this specification is currently on-hold in favour of developing SED-ML to meet the requirements originally being addressed via the approach specified below.
This version:
Last version:
Latest version:
Authors:
Abstract
CellML provides a mechanism to describe mathematical models. While CellML describes the mathematics used by a model, it does not describe the details of any particular simulation being run. As such, a CellML model does not in itself provide sufficient information to reproduce experimental results in a completely automated fashion.
It would be extremely useful if authors could provide information in a machine readable format, not just about their model, but also about the particular simulation they have run. In this way, it becomes possible to reproduce any simulation results obtained by the author simply by loading the information into a simulation tool and asking it to re-run the simulation.
Such data fits naturally into a CellML model, as metadata. CellML metadata is expressed in Resource Description Format(RDF). This specification provides an RDF based language for describing simulations. As RDF is an open-world framework, such RDF data may be embedded in a CellML document, or alternatively, could be provided by a third party in a separate document.
Status of this document
This document is a discussion draft version of the CellML simulation metadata specification, and has not been endorsed by the CellML team or members of the CellML community. Comments are solicited and should be sent to cellml-discussion@cellml.org.
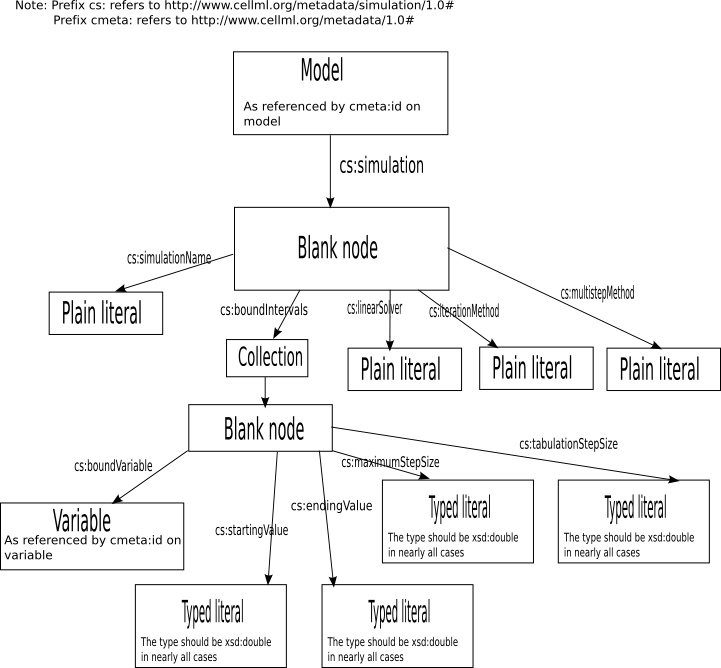
Graphical summary of core features(non-normative)
Prefix
The URI http://www.cellml.org/metadata/simulation/1.0# is used as the prefix for all RDF predicates defined in this specification. When describing documents in RDF/XML, it is recommended that the prefix cs be bound to this URI. However, processing software which parses CellML simulation metadata in RDF/XML form must not assume that this prefix will be used.
Within this document, predicate resources are referred to using the notation of qualified names in RDF/XML, and assuming that the cs prefix is bound to "http://www.cellml.org/metadata/simulation/1.0#". For example, cs:simulation refers to the resource "http://www.cellml.org/metadata/simulation/1.0#simulation".
Within this document, it is assumed that the prefix cmeta is bound to the XML namespace "http://www.cellml.org/metadata#1.0#". However, processing software must not assume that this prefix will always be used.
Specification of simulations
As described in the CellML metadata specification, the CellML model is associated with the RDF resource with fragment identifier matching the cmeta:id on the model element and the URI base equal to the CellML document URI.
Every CellML model may have zero or more simulations associated with them. Such simulations are specified by an arc from the model (subject) to a node(object), with predicate cs:simulation. As one model can have more than one simulation associated with it, it is not a contradiction for two or more such arcs to exist. Where CellML processing software discovers more than one such arc, it may prompt the user to select one, or it may choose an arc arbitrarily, or it may process two or more arcs.
Throughout this document, the object node of the arc just described is referred to as the "simulation node".
The simulation node shall be closed-world node with respect to predicates defined in this specification — that is, it shall be completely specified within the XML file in which it is defined. If a user wishes to add additional details defined in this specification to a different XML document, they must not attempt to define new arcs with the simulation node as the subject. Instead, they should define a new simulation with the additional information. However, it is acceptable for predicates which annotate the simulation(for example, some future specification containing curation information), rather than define it, to be specified in a different XML file.
All simulations shall have a name. This name shall be specified by an arc from the simulation node to a plain literal(containing the name) with predicate cs:simulationName.
Simulations may optionally also have information about the algorithms to be used for the simulation. There may be either zero or one arcs from the simulation node to an object, for each of the predicates listed below:
- cs:linearSolver : The target of this predicate should be a plain literal, containing the name of the linear solver to use. Possible values include direct (to use a direct linear solver), direct-dense and direct-banded (as before, but hinting whether to use a dense or banded Jacobian), generalised-minimal-residual (for GMRES / SPGMR type iterative methods), biconjugate-gradiant (for BiCG / SPBCG) and quasi-minimal-residual (or transpose-free-quasi-minimal-residual) for QMR or TFQMR methods. Implementors may add new values. If an implementation cannot deal with different linear solvers, or it doesn't recognise the value present, it should fall back on a direct method.
- cs:iterationMethod: The target of this predicate should be a plain literal, containing the name of the iteration method to use. An arc with this predicate should only be present if the target of the cs:linearSolver is an iterative method. Possible values include newton (to use Newton iterations) or krylov-subspace (to use a Krylov subspace iteration method). Implementors may add new values. If an implementation cannot deal with the iteration method specified, it should fall back onto the Newton solver.
- cs:multistepMethod: The target of this predicate should be a plain literal, containing the name of the multi-step method to use. Possible values include adams-moulton (to use an implicit Adams-Moulton method of arbitrary order. The order can optionally be appended to get a value like adams-moulton-2), embedded-runge-kutta-2 (To use an embedded second order Runge-Kutta method), embedded-runge-kutta-4, embedded-runge-kutta-4-estimate-5 (to use a fourth order embedded Runge-Kutta method, with a fifth order error estimate), runge-kutta-cash-karp-4-estimate-5 (to use a fourth order Runge-Kutta Cash-Karp method, with a fifth order error estimate), runge-kutta-fehlberg-4-estimate-5 (as for the previous, but using an RKF method), runge-kutta-prince-dormand-8-estimate-9 (8th order RKPD, with a 9th order error estimate), implicit-runge-kutta-2 (an implicit second order Runge Kutta method), implicit-runge-kutta-4 (an implicit fourth order Runge-Kutta methd), bulirsch-stoer-bader-deuflhard (the Bulirsch Stoer method of Bader and Deuflhard), gear-1 (the first order backwards differential formula of Gear), gear-2 (the second order backwards differential formula of Gear). Implementors may add new values. If an implementation cannot deal with the stepping method specified, it may fall back on the method of its choice.
Specification of bound variable ranges
Simulations based on differential equations are generally run across a range of bound variable values. In order to reproduce simulation results, it is important to know what bound variable intervals are involved.
The document defining the simulation node must have exactly one arc with predicate cs:boundIntervals. The object node of this arc shall be of type RDF:List, and shall follow the standard semantics for a collection. It shall be defined as a linear list(with no branching structure), and shall be explicitly terminated with RDF:nil.
Each member of the collection shall be a node, which is referred to as a bound variable node throughout this specification. Each bound variable node shall be closed-world node with respect to predicates defined in this specification — that is, it shall be completely specified within the XML file in which it is defined. Please refer to the section on the simulation node for futher discussion of this.
The document defining the bound variable nodes must have an arc from the bound variable node to a resource associated with a variable in the CellML model, under the normal rules for asscoiating an element to a resource using cmeta:id. This arc shall have the predicate cs:boundVariable.
In addition, the document must have an arc from the bound variable node to a typed literal with predicate cs:startingValue. The document must also have an arc from the bound variable node to a typed literal with predicate cs:endingValue. These two literal define the starting and ending values of the interval on which the model is to be integrated. These literals should be of type xsd:double, unless xsd:double cannot represent the type of the bound variable. CellML processing software is not required to support any type other than xsd:double.
The document may contain at most one arc from each bound variable node subject with the predicate cs:maximumStepSize. If present, this value specifies the upper limit on any steps which may be taken. These literals should be of type xsd:double, unless xsd:double cannot represent the type of the bound variable. CellML processing software is not required to support any type other than xsd:double.
All literals of type xsd:double defined in this section are expressed in terms if the bound variable units. The bound variable units are the units on the target bound variable. The target bound variable is the bound connected variable with no in interfaces. The set of bound connected variables for the bound variable anonymous node contains:
- The variable with the cmeta:id referenced by the cs:boundVariable target, and,
- All variables connected (by a CellML connection) to a bound connected variable.
The document may contain at most one arc from each bound variable node subject with the predicate cs:tabulationStepSize. If present, this value gives a hint at the distance between values which should be reported back to the user. These literals should be of type xsd:double, unless xsd:double cannot represent the type of the bound variable. CellML processing software is not required to support any type other than xsd:double. Developers of some classes of CellML processing software may find that this information is irrelevant, or does not make sense for the type of output produced by that software. This software may silently ignore the cs:tabulationStepSize arc.
Appendix A: Sample model with simulation metadata (non-normative)
<?xml version="1.0" encoding="iso-8859-1"?>
<!--
* Version: MPL 1.1/GPL 2.0/LGPL 2.1
*
* The contents of this file are subject to the Mozilla Public License
* Version 1.1 (the "License"); you may not use this file except in
* compliance with the License. You may obtain a copy of the License at
* http://www.mozilla.org/MPL/
*
* Software distributed under the License is distributed on an "AS IS" basis,
* WITHOUT WARRANTY OF ANY KIND, either express or implied. See the License
* for the specific language governing rights and limitations under the
* License.
*
* The Original Code is the mozCellML code.
*
* The Initial Developer of the Original Code is
* The Bioengineering Institute, University of Auckland.
* Portions created by the Initial Developer are Copyright (C) 2004
* the Initial Developer. All Rights Reserved.
*
* Contributor(s):
* Andrew Miller <ak.miller@auckland.ac.nz> (original developer)
*
* Alternatively, the contents of this file may be used under the terms of
* either the GNU General Public License Version 2 or later (the "GPL"), or
* the GNU Lesser General Public License Version 2.1 or later (the "LGPL"),
* in which case the provisions of the GPL or the LGPL are applicable instead
* of those above. If you wish to allow use of your version of this file only
* under the terms of either the GPL or the LGPL, and not to allow others to
* use your version of this file under the terms of the MPL, indicate your
* decision by deleting the provisions above and replace them with the notice
* and other provisions required by the GPL or the LGPL. If you do not delete
* the provisions above, a recipient may use your version of this file under
* the terms of any one of the MPL, the GPL or the LGPL.
*
-->
<model
name="CoupledPendulum_version01"
xmlns="http://www.cellml.org/cellml/1.0#"
xmlns:cmeta="http://www.cellml.org/metadata/1.0#"
xmlns:cellml="http://www.cellml.org/cellml/1.0#"
cmeta:id="CoupledPendulum_version01">
<rdf:RDF
xmlns:rdf="http://www.w3.org/1999/02/22-rdf-syntax-ns#"
xmlns:dc="http://purl.org/dc/elements/1.1/"
xmlns:cmeta="http://www.cellml.org/metadata/1.0#"
xmlns:dcterms="http://purl.org/dc/terms/"
xmlns:vCard="http://www.w3.org/2001/vcard-rdf/3.0#"
xmlns:bqs="http://www.cellml.org/bqs/1.0#"
xmlns:cs="http://www.cellml.org/metadata/simulation/1.0#">
<rdf:Description rdf:about="">
<dc:creator rdf:parseType="Resource">
<vCard:N rdf:parseType="Resource">
<vCard:Family>Miller</vCard:Family>
<vCard:Given>Andrew</vCard:Given>
<vCard:Other>Keith</vCard:Other>
</vCard:N>
<vCard:EMAIL rdf:parseType="Resource">
<rdf:value>ak.miller@auckland.ac.nz</rdf:value>
<rdf:type rdf:resource="http://imc.org/vCard/3.0#internet" />
</vCard:EMAIL>
<vCard:ORG rdf:parseType="Resource">
<vCard:Orgname>The University of Auckland</vCard:Orgname>
<vCard:Orgunit>The Bioengineering Institute</vCard:Orgunit>
</vCard:ORG>
</dc:creator>
<dcterms:created rdf:parseType="Resource">
<dcterms:W3CDTF>2004-12-09</dcterms:W3CDTF>
</dcterms:created>
<dcterms:modified rdf:parseType="Resource">
<dcterms:W3CDTF>2006-08-14</dcterms:W3CDTF>
</dcterms:modified>
<dc:publisher>
The University of Auckland, The Bioengineering Institute
</dc:publisher>
</rdf:Description>
<rdf:Description rdf:about="#CoupledPendulum_version01">
<dc:title>Coupled Pendulum Model</dc:title>
<bqs:WebResource rdf:parseType="Resource">
<bqs:url>http://www.cs.wisc.edu/~hasti/cs310/notes/ODE1/ODE1Notes.html</bqs:url>
</bqs:WebResource>
<cs:simulation rdf:parseType="Resource">
<cs:simulationName>SwingFor100s</cs:simulationName>
<cs:multistepMethod>implicit-runge-kutta-2</cs:multistepMethod>
<cs:linearSolver>direct</cs:linearSolver>
<cs:variablesImportantInSimulation rdf:parseType="Collection">
<rdf:Description rdf:about="#time" />
<rdf:Description rdf:about="#a_angle" />
<rdf:Description rdf:about="#b_angle" />
</cs:variablesImportantInSimulation>
<cs:boundIntervals rdf:parseType="Collection">
<rdf:Description>
<cs:boundVariable><rdf:Description rdf:about="#time" /></cs:boundVariable>
<cs:maximumStepSize rdf:datatype="http://www.w3.org/2001/XMLSchema#double">1</cs:maximumStepSize>
<cs:tabulationStepSize rdf:datatype="http://www.w3.org/2001/XMLSchema#double">0.1</cs:tabulationStepSize>
<cs:startingValue rdf:datatype="http://www.w3.org/2001/XMLSchema#double">0</cs:startingValue>
<cs:endingValue rdf:datatype="http://www.w3.org/2001/XMLSchema#double">100</cs:endingValue>
</rdf:Description>
</cs:boundIntervals>
</cs:simulation>
</rdf:Description>
</rdf:RDF>
<units name="rad_per_s">
<unit units="radian" />
<unit units="second" exponent="-1" />
</units>
<units name="rad2">
<unit units="radian" exponent="2" />
</units>
<component name="environment">
<variable name="time" public_interface="out" units="second" cmeta:id="time" />
</component>
<component name="PendulumUpperSegment">
<variable name="a" units="radian" public_interface="out" cmeta:id="a_angle" initial_value="1"/>
<variable name="a_angular_velocity" units="rad_per_s"
public_interface="out" initial_value="0"/>
<variable name="time" units="second" public_interface="in"/>
<math xmlns="http://www.w3.org/1998/Math/MathML">
<apply>
<eq/>
<apply>
<diff/>
<bvar><ci>time</ci></bvar>
<ci>a</ci>
</apply>
<ci>a_angular_velocity</ci>
</apply>
</math>
</component>
<component name="PendulumLowerSegment">
<variable name="b" initial_value="1" units="radian" public_interface="out" cmeta:id="b_angle" />
<variable name="b_angular_velocity" initial_value="0"
units="rad_per_s" public_interface="out"/>
<variable name="time" units="second" public_interface="in"/>
<math xmlns="http://www.w3.org/1998/Math/MathML">
<apply>
<eq/>
<apply>
<diff/>
<bvar><ci>time</ci></bvar>
<ci>b</ci>
</apply>
<ci>b_angular_velocity</ci>
</apply>
</math>
</component>
<component name="Pendulum">
<variable name="a" private_interface="in" public_interface="out" units="radian"/>
<variable name="b" private_interface="in" public_interface="out" units="radian" initial_value="1"/>
<variable name="a_angular_velocity" private_interface="in" units="rad_per_s"/>
<variable name="b_angular_velocity" private_interface="in" units="rad_per_s"/>
<variable name="time" public_interface="in" private_interface="out" units="second"/>
<math xmlns="http://www.w3.org/1998/Math/MathML">
<apply>
<eq/>
<apply>
<diff/>
<bvar><ci>time</ci></bvar>
<ci>a_angular_velocity</ci>
</apply>
<apply>
<plus/>
<apply>
<times/>
<cn cellml:units="dimensionless">-2.0</cn>
<ci>a</ci>
</apply>
<ci>b</ci>
</apply>
</apply>
<apply>
<eq/>
<apply>
<diff/>
<bvar><ci>time</ci></bvar>
<ci>b_angular_velocity</ci>
</apply>
<apply>
<minus/>
<apply>
<times/>
<cn cellml:units="dimensionless">2.0</cn>
<ci>a</ci>
</apply>
<apply>
<times/>
<cn cellml:units="dimensionless">2</cn>
<ci>b</ci>
</apply>
</apply>
</apply>
</math>
</component>
<component name="analytic_solution">
<variable name="a_" units="radian" public_interface="out"/>
<variable name="b_" units="radian" public_interface="out"/>
<variable name="time" units="second" public_interface="in"/>
<math xmlns="http://www.w3.org/1998/Math/MathML">
<apply><eq/>
<ci>a_</ci>
<apply><plus/>
<apply><times/>
<cn cellml:units="dimensionless">0.5</cn>
<apply><cos/>
<apply><times/>
<!-- sqrt(2 - sqrt(2)) -->
<cn cellml:units="dimensionless">0.765366864730179</cn>
<ci>time</ci>
</apply>
</apply>
</apply>
<apply><times/>
<cn cellml:units="dimensionless">0.25</cn>
<!-- sqrt(2) -->
<cn cellml:units="dimensionless">1.41421356237310</cn>
<apply><cos/>
<apply><times/>
<!-- sqrt(2 - sqrt(2)) -->
<cn cellml:units="dimensionless">0.765366864730179</cn>
<ci>time</ci>
</apply>
</apply>
</apply>
<apply><times/>
<cn cellml:units="dimensionless">0.5</cn>
<apply><cos/>
<apply><times/>
<!-- sqrt(2 + sqrt(2)) -->
<cn cellml:units="dimensionless">1.84775906502257</cn>
<ci>time</ci>
</apply>
</apply>
</apply>
<apply><times/>
<cn cellml:units="dimensionless">-0.25</cn>
<!-- sqrt(2) -->
<cn cellml:units="dimensionless">1.41421356237310</cn>
<apply><cos/>
<apply><times/>
<!-- sqrt(2 + sqrt(2)) -->
<cn cellml:units="dimensionless">1.84775906502257</cn>
<ci>time</ci>
</apply>
</apply>
</apply>
</apply>
</apply>
<apply><eq/>
<ci>b_</ci>
<apply><plus/>
<apply><times/>
<cn cellml:units="dimensionless">0.5</cn>
<cn cellml:units="dimensionless">1.41421356237310</cn>
<apply><cos/>
<apply><times/>
<!-- sqrt(2 - sqrt(2)) -->
<cn cellml:units="dimensionless">0.765366864730179</cn>
<ci>time</ci>
</apply>
</apply>
</apply>
<apply><times/>
<cn cellml:units="dimensionless">0.5</cn>
<apply><cos/>
<apply><times/>
<!-- sqrt(2 - sqrt(2)) -->
<cn cellml:units="dimensionless">0.765366864730179</cn>
<ci>time</ci>
</apply>
</apply>
</apply>
<apply><times/>
<cn cellml:units="dimensionless">-0.5</cn>
<!-- sqrt(2) -->
<cn cellml:units="dimensionless">1.41421356237310</cn>
<apply><cos/>
<apply><times/>
<!-- sqrt(2 + sqrt(2)) -->
<cn cellml:units="dimensionless">1.84775906502257</cn>
<ci>time</ci>
</apply>
</apply>
</apply>
<apply><times/>
<cn cellml:units="dimensionless">0.5</cn>
<apply><cos/>
<apply><times/>
<!-- sqrt(2 + sqrt(2)) -->
<cn cellml:units="dimensionless">1.84775906502257</cn>
<ci>time</ci>
</apply>
</apply>
</apply>
</apply>
</apply>
</math>
</component>
<component name="errors">
<variable name="a" public_interface="in" units="radian"/>
<variable name="a_" public_interface="in" units="radian"/>
<variable name="b" public_interface="in" units="radian"/>
<variable name="b_" public_interface="in" units="radian"/>
<variable name="error_A" units="rad2"/>
<variable name="error_B" units="rad2"/>
<variable name="RSS" units="rad2"/>
<math xmlns="http://www.w3.org/1998/Math/MathML">
<apply><eq/>
<ci>error_A</ci>
<apply><power/>
<apply><minus/>
<ci>a</ci>
<ci>a_</ci>
</apply>
<cn cellml:units="dimensionless">2</cn>
</apply>
</apply>
<apply><eq/>
<ci>error_B</ci>
<apply><power/>
<apply><minus/>
<ci>b</ci>
<ci>b_</ci>
</apply>
<cn cellml:units="dimensionless">2</cn>
</apply>
</apply>
<apply><eq/>
<ci>RSS</ci>
<apply><plus/>
<ci>error_A</ci>
<ci>error_B</ci>
</apply>
</apply>
</math>
</component>
<connection>
<map_components component_1="environment" component_2="Pendulum"/>
<map_variables variable_1="time" variable_2="time"/>
</connection>
<connection>
<map_components component_1="environment" component_2="analytic_solution"/>
<map_variables variable_1="time" variable_2="time"/>
</connection>
<connection>
<map_components component_1="Pendulum" component_2="PendulumUpperSegment"/>
<map_variables variable_1="time" variable_2="time"/>
<map_variables variable_1="a" variable_2="a"/>
<map_variables variable_1="a_angular_velocity" variable_2="a_angular_velocity"/>
</connection>
<connection>
<map_components component_1="Pendulum" component_2="PendulumLowerSegment"/>
<map_variables variable_1="time" variable_2="time"/>
<map_variables variable_1="b" variable_2="b"/>
<map_variables variable_1="b_angular_velocity" variable_2="b_angular_velocity"/>
</connection>
<connection>
<map_components component_1="errors" component_2="analytic_solution"/>
<map_variables variable_1="a_" variable_2="a_"/>
<map_variables variable_1="b_" variable_2="b_"/>
</connection>
<connection>
<map_components component_1="errors" component_2="Pendulum"/>
<map_variables variable_1="a" variable_2="a"/>
<map_variables variable_1="b" variable_2="b"/>
</connection>
<group>
<relationship_ref relationship="encapsulation"/>
<component_ref component="Pendulum">
<component_ref component="PendulumUpperSegment"/>
<component_ref component="PendulumLowerSegment"/>
</component_ref>
</group>
</model>

