basic_ep_model_doc
Simple Membrane / Electrophysiological Models
Table of Contents
This document presents a simple model of a cell, demonstrating the basic concepts in defining an electrophysiological model using CellML. This model is not intended as anything more than an example and is not supposed to be realistic.
You might find it helpful to refer to a full printout of the CellML document that makes up this example as you progress through this documentation. The various forms available online are presented in the section “Download This Model”.
The model in this example consists of a one-compartment cell, with the interior of the cell separated from the extracellular space by a membrane, as shown in Figure 1. There are two channels in the membrane, which allow sodium and calcium ions to flow from the extracellular subspace into the intracellular subspace. All of the variables in the model are scalars that vary only with time. This type of model is often referred to as a “lumped parameter” model.

In CellML, models are thought of as connected networks of discrete components. These components may correspond to physiologically separated regions or may be convenient modelling abstractions. The CellML network rendering of the simple model introduced in Figure 1 is shown in Figure 2. The different shapes in the diagram are explained in the notation guide. The model consists of three components representing physiologically separated regions (the intracellular space, the cellular membrane and the extracellular space) and one component representing the environment, providing a convenient container to store global variables such as time. The environment component is shown in Figure 2 to illustrate how these variables are handled in CellML. However, it is typically omitted from the CellML network rendering, as it will always exist, and it is usually connected to every other component.

Figure 2 also shows where variables are declared and how they are passed around between components in the model. The variables are displayed next to the component in which they are declared, alongside the line that represents the connection to another component, which imports and uses their values. Variables that represent the concentrations of calcium and sodium are declared in both the intra- and extra-cellular subspaces and passed into the membrane for use in the calculation of the fluxes of calcium and sodium ions. The variables that represent these currents are declared in the membrane and passed back into the intra- and extra-cellular subspaces, in which they are used by the equations that update the ion concentrations in the two subspaces. The rate constant variables declared in the cell membrane are not visible outside of that component and are therefore not shown in this rendering.
The root element of the CellML document for this simple electrophysiological model is the <model> element shown in Figure 3. (A CellML document is an XML document that contains the CellML description of a model. Those not familiar with XML may want to consult the quick introduction to XML for help with understanding XML terms.) The <model> element has a name attribute that allows this model to be unambiguously referenced by other models. For instance, this would be necessary if this model were to be combined with other models or partial models to create a larger model.
<model> element shown above.
Two namespaces are also declared on the <model> element. The first sets the default namespace for the <model> element and all elements contained within the <model> element to the CellML 1.0 namespace. The second namespace is again the CellML 1.0 namespace, but this time mapped to the cellml prefix. This declaration has document-wide scope, so the cellml prefix may be used anywhere to move an element or attribute into the CellML namespace. The declaration of the CellML namespace as both the default namespace and as a namespace mapped to the cellml prefix is recommended practice for any <model> element. This simplifies the use of CellML elements and attributes inside MathML. For instance, a cellml:units attribute (described in the section “Math”) can be added to <cn> elements without having to re-declare the CellML namespace with each occurrence.
For a more in-depth explanation of XML namespaces in CellML, see Section 2.2.2 of the CellML specification.
The CellML specification defines a standard dictionary of units that can be used in CellML models without further definition. This dictionary consists of the base SI units (as defined by the NIST) and some additional units commonly found in the types of biological models likely to be defined using CellML.
A modeller who wishes to use units not declared in the standard dictionary can define additional units in the CellML model document. The units definitions that are needed by the example model are shown in Figure 4. An identifier is associated with each units definition, which is composed of a combination of base units from the CellML standard dictionary and units that have been previously defined in the current model. The identifiers can then be referenced in the units attribute on <variable> and <cn> elements (<cn> elements enclose bare numbers in MathML). CellML processing software may choose to use these units definitions to verify the consistency of units across connections (inserting scale-factors, if appropriate) and check the dimensions of equations, as described in Section 5.2.6 and Section 5.2.7 of the CellML specification, respectively.
<unit> elements gives a new units definition.
The units definitions are declared with a set of <units> elements. Each <units> element must have a name attribute, which declares the identifier that may be used to refer to the unit in the rest of the document.
Each <units> element may contain one or more <unit> elements. The units defined by the <units> element is the combination of the contents of these <unit> elements. Section 5.2.2 of the CellML specification explains how the attributes for each <unit> element contribute to a new units definition.
Looking at the units that are defined in Figure 4, we see that the first one is a millimole/litre, which will be represented by the identifier "concentration_units". The second unit definition defines a unit for the change in concentration over time, millimoles/litre-second, which is given the name "flux_units". The third <units> element defines inverse seconds and is given the name "rate_constant" .
The first <component> element defined in the example model is called environment. The complete definition is given in Figure 5. This component does not correspond to a physical compartment in a cell, but rather is an abstract container used to define variables that are used throughout the model.
The <component> element must have a single name attribute, which must be unique across the model. The value of the name attribute is used to reference the component in connections and groups.
environment component represents a convenient abstract container, which is used to define the independent variable time separately from the rest of the model.
In our simple model, the only truly global variable is the independent variable time. In CellML models, no variables are given any kind of precedence or implicitly assumed to exist. This makes the model definition more robust and model components more re-usable. Therefore, it is necessary to declare variables to represent time and space if they are needed in a model, but it is not necessary to indicate that a variable like time is an independent variable, as this can be determined from analysis of the differential equations. Theoretically, you could call the variable that represents time A1 and use the names t and time to represent other concepts in your model. However, in practice, it would be unwise to use the names time and t to represent anything other than time, or the names x, y, or z to represent anything other than space. Doing so would make it more difficult for other model authors to re-use your work.
The <variable> element is explained in the section “The intra_cellular_space component”.
The environment component is extremely simple. The second <component> element, which corresponds to the intra-cellular compartment, better demonstrates the functionality and structure of a <component> element. The definition of this component is shown in Figure 6, with the contents of the <math> element omitted (math is discussed in the section “Math”).
<component> element corresponding to the intra-cellular compartment in the example model. The contents of the <math> element have been left out (math is discussed in the section “Math”).
Variables may only be declared inside a <component> element. A variable is not visible to other components unless it is explicitly exposed using the public_interface or private_interface attributes. The private_interface attribute is used in conjunction with encapsulation, which is discussed in a separate example. The allowed usage of each variable in the current model is determined by the value of its public_interface attribute. Variables with a public_interface value of "out" may have their value modified in the current component, and their value may be mapped to variables in other components with a public_interface value of "in". The intra-cellular component contains two such variables: the concentrations of sodium and calcium ions, declared with identifiers Na and Ca, respectively.
Variables with a public_interface value of "in" obtain their value from mappings to variables declared in other components with a public_interface value of "out". Their value may not be modified in the current component. The intra-cellular component declares three such variables: the independent variable time and the fluxes of sodium and calcium ions across the membrane, denoted by I_Na and I_Ca, respectively.
Each <variable> element must have a units attribute. The value of this attribute must correspond to a units definition in the standard CellML dictionary or to units defined within the current component or model.
The definition of the cell_membrane component is shown in Figure 7, with the contents of the <math> element again omitted. The membrane component introduces a third kind of variable: one without a public_interface attribute. Variables with a public_interface value of "none" or without a public_interface attribute may be modified in the current component, but are not visible to other components. Such variables can be thought of as private to the current component, and are generally convenient for storing the values of intermediate values, or constants that the model user might want to modify. The rate constants for the diffusion of sodium and calcium ions across the membrane fit in the latter category and are denoted by v_Na and v_Ca, respectively.
Single-valued variables with a public_interface attribute value of "out" or "none" may have their starting value set using the initial_value attribute on the <variable> element, as shown for the rate constants in the membrane component. The CellML specification (Section 3.2) states that the value of the initial_value attribute corresponds to the value of the variable when all independent variables (the variables with respect to which differentiation or integration occurs) have a value of 0.0. Since time is the only independent variable in this model, the initial values of v_Na and v_Ca are initial conditions with respect to time. Initial values are typically not specified for the majority of variables in a model as these values can be set by simulation software from external configuration files or user input.
Section 3.2 of the CellML specification has more on the <component> and <variable> elements.
Figure 8 shows the <math> element and its contents from the definition of the extra-cellular component. The <math> element contains the governing equations for the conservation of sodium and calcium ion concentrations in the extra-cellular compartment. The second of these equations has been omitted from Figure 8 for brevity. The equations are defined using MathML, a XML vocabulary being developed by the World Wide Web Consortium.
<math> element from the extra-cellular component. This equation governs the behaviour of the extra-cellular sodium concentration. An identical equation governing the behaviour of the intra-celluar calcium concentration was omitted.The equation defined in Figure 8 is:
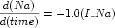
The default namespace for the <math> element and all of its children elements is set to the MathML namespace as defined in the MathML 2.0 specification, overriding the default namespace declaration on the <model> element. The single number that occurs in the equation is assigned units by adding a cellml:units attribute to the <cn> element. This attribute is not part of the MathML specification. It is placed in the CellML namespace using the cellml prefix declared on the <model> element (as shown in Figure 3). Any number occurring in a MathML block should be placed in a <cn> element and assigned units in this manner. This supports basic consistency checking of the equation.
<connection> elements are used to define the mappings between variables with a public_interface value of "out" in one component to variables with a public_interface value of "in" in another component. Figure 9 shows the <connection> element that is used to specify the mappings between variables in the intra-cellular and membrane components. Only one connection can be created between any two components in a model, so mappings in both directions must be stored together within the same <connection> element.
Each <connection> element must contain a single <map_components> element which references (in any order) two components contained in the current <model> element. The variables that are being mapped between the two components are listed successively in separate <map_variables> elements. The value of the variable_1 attribute must be the name of a <variable> element contained in the <component> referenced by the component_1 attribute on the <map_component> element, and the value of the variable_2 attribute must correspond to a variable in component_2.
The direction of each mapping is determined by the value of the public_interface attributes on the two variables: the value is always passed from the variable with a public_interface of "out" to the variable with a public_interface of "in". In this case, the concentration of sodium from the intra-cellular component denoted by Na is mapped to the Na_i variable in the membrane. The sodium flux I_Na is passed in the other direction.
The CellML description of this model is available in a number of formats. If you have your browser set up to view text files served with the “text/xml” MIME type, then you can have a look at the XML file here. If not, you can save that file to disk by shift-clicking on the preceding link. A “pretty-printed” browsable HTML version of the XML file is available here
—
note that you cannot download and save this version for later viewing since it makes use of stylesheets for formatting. If you wish to save or print out the “pretty-printed” version of the XML, a PDF version is also available here.
Here are those links again:
basic_ep_model.xml — the raw XML.
basic_ep_model.html — an HTML version for browsing online.
basic_ep_model.pdf — a PDF version suitable for printing.
basic_ep_model_maths.pdf — a PDF of the equations described in the model generated directly from the CellML description using the MathML Renderer.

