Using the import feature of CellML 1.1 to reuse existing models
Describing heart cells
Jonna Terkildsen completed her Masters thesis at the Auckland Bioengineering Institute, The University of Auckland and has recently started her PhD. Jonna has created a number of case-studies demonstrating the use of the import feature of CellML 1.1. One such case study is the coupling of three existing published mathematical models of rat heart cell function (an electrophysiology model (Pandit et al., Biophysical Journal 81:3029-3051, 2001), a model of cellular calcium dynamics (Hinch et al., Biophysical Journal 87:3723-3736, 2004) and a mechanics model (Niederer et al., Biophysical Journal 90:1697-1722, 2006)) to produce a single, detailed, accurate electomechanic cell model (Terkildsen et al., Experimental Physiology 93:919-929, 2008).
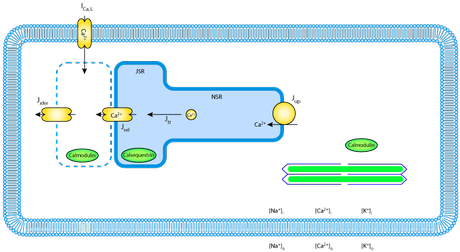
Schematic of the Pandit 2001 model.
The pros and the cons of reusing existing models
Using the import feature of CellML 1.1, Jonna was able to couple the models together – and in doing so demonstrated the advantages and disadvantages associated with the process of model reuse. On the one hand she was able to use existing work and develop it further, while on the other the process of combining models required a degree of adaptation – such as units conversion, and maintaining consistency between variable names.
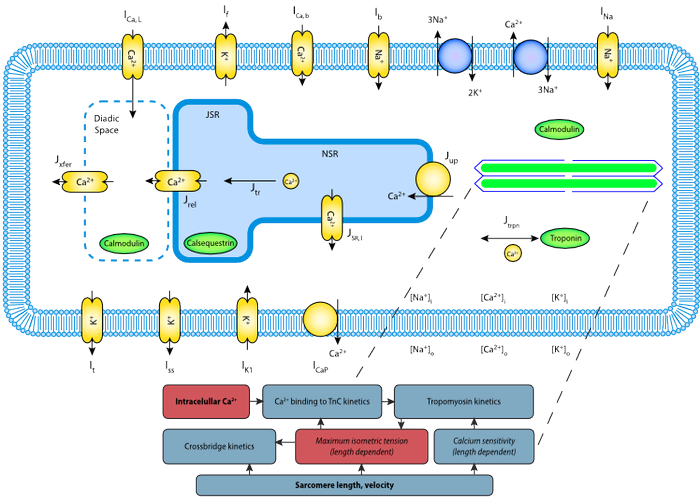
Schematic of the Terkildsen 2008 model, a coupling of three different models using CellML 1.1 import functions.
Applying similar techniques to describe cardiovascular circulation
Jonna has also implemented the 1992 Guyton Cardiovascular Circulation model in CellML – both CellML 1.0 and CellML 1.1. This large model was implemented as a central Circulation Dynamics model, the behaviour of which is influenced by 20 modules. These modules each represent a separate physiological subsystem (for example, the dynamics of the kidney, electrolytes and cell water, thirst and drinking, hormone regulation, autonomic regulation, cardiovascular system) and these feedback on the central circulation model. The 20 modules were coded up as separate CellML models and also combined into a complete model.