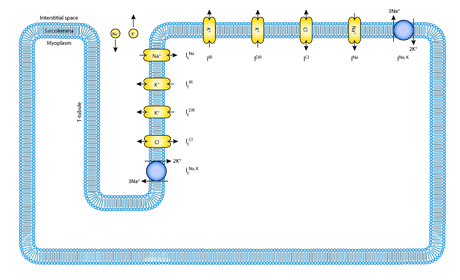
Using CellML to embed muscle cell models in a multiscale tissue network

Using a suite of tools
To achieve this John translated the model, which was originally developed in MATLAB, into CellML using the Matlab2CellML converter. Further editing was carried out in a text editor, and the validity of the final model was tested using the simulation tools JSim and CMISS. In this case, the main advantage of CellML over the original MATLAB was that it allowed John to embed his cell model within a larger tissue framework.
Schematic diagram of John's muscle model
From skeletal muscle to smooth muscle in the gut
Since completing his PhD, John has moved on to study the gut. He is now working with an existing CellML model of pacemaker activity in the interstitial cells of Cajal (ICC), developed by his colleague Richard Faville. Using CMISS, John has embedded this cell model in a larger tissue framework to more accurately study slow-wave activity in the gut.